Selective Inhibitors of Histone Methyltransferase DOT1L: Design, Synthesis, and Crystallographic Studies.
Yao, Y., Chen, P., Diao, J., Cheng, G., Deng, L., Anglin, J.L., Prasad, B.V., Song, Y.(2011) J Am Chem Soc 133: 16746-16749
- PubMed: 21936531
- DOI: https://doi.org/10.1021/ja206312b
- Primary Citation of Related Structures:
3SR4 - PubMed Abstract:
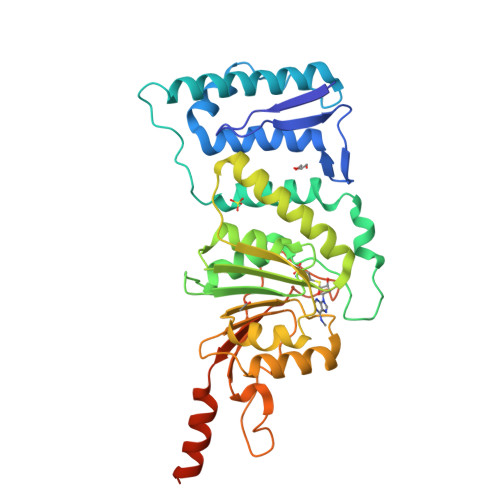
Histone H3-lysine79 (H3K79) methyltransferase DOT1L plays critical roles in normal cell differentiation as well as initiation of acute leukemia. We used structure- and mechanism-based design to discover several potent inhibitors of DOT1L with IC(50) values as low as 38 nM. These inhibitors exhibit only weak or no activities against four other representative histone lysine and arginine methyltransferases, G9a, SUV39H1, PRMT1 and CARM1. The X-ray crystal structure of a DOT1L-inhibitor complex reveals that the N6-methyl group of the inhibitor, located favorably in a predominantly hydrophobic cavity of DOT1L, provides the observed high selectivity. Structural analysis shows that it will disrupt at least one H-bond and/or have steric repulsion for other histone methyltransferases. These compounds represent novel chemical probes for biological function studies of DOT1L in health and disease.
Organizational Affiliation:
Department of Pharmacology, Baylor College of Medicine, 1 Baylor Plaza, Houston, Texas 77030, USA.